Week 2: Designing Synthetic BIology Systems
What I cannot create, I do not understand
--- Richard Feynman
What about this week on Designing Synthetic Biology Systems?
George Church deeped dived into three SynBio projects: Multiplex synthetic libraries & selections, Recoding: resistance to all viruses and Editing repeats.
What gene you want to edit?
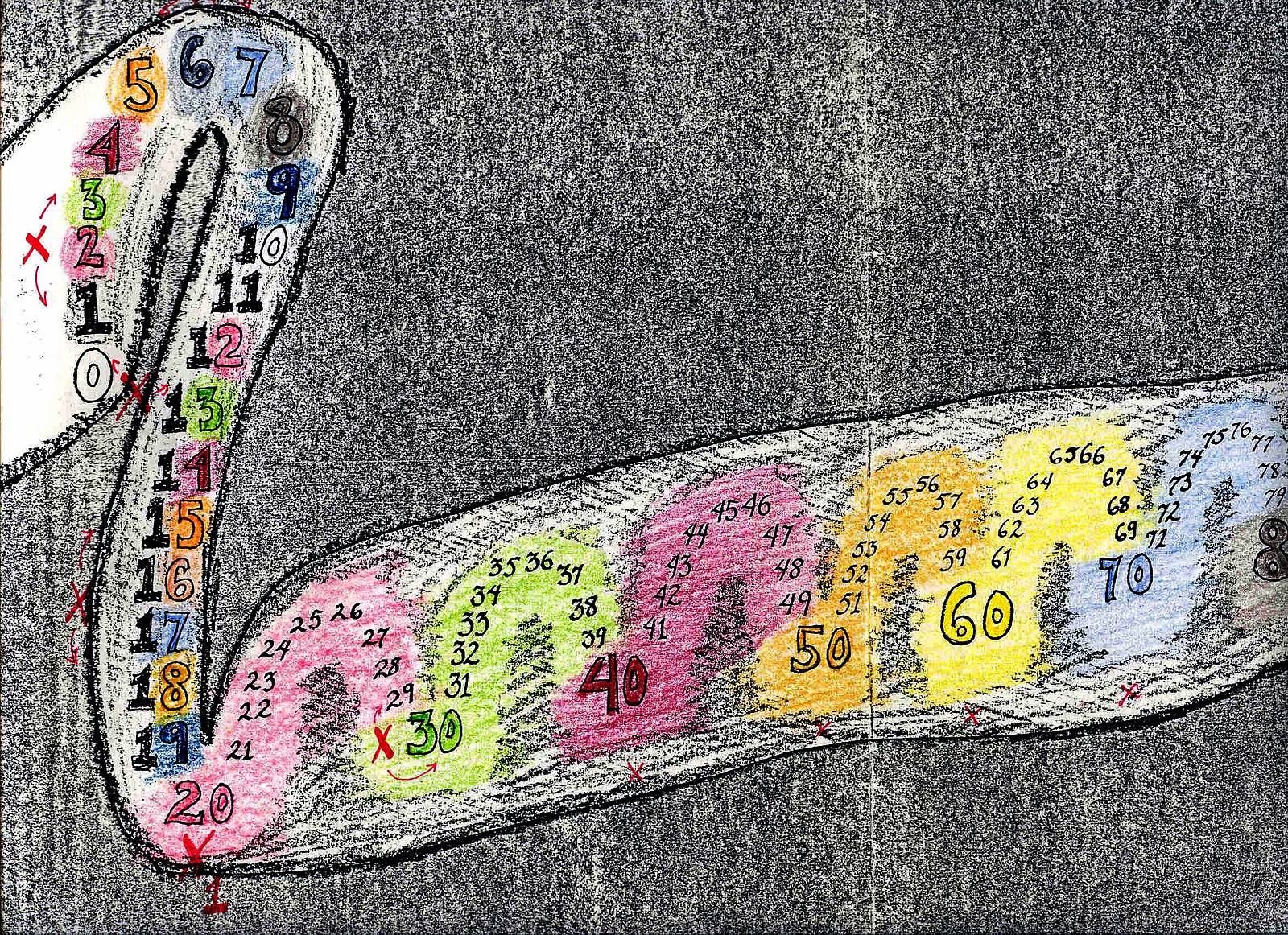
from Wikipedia
Genes related to superpower of sensory
Some people are able to connect different sensory such as hearing a color, which is called Synesthesia. Athough little do we know the exactly genetic mechanism behind this, one publication on PNAS showed rare variants maybe contribute to one kinds of sound-color synesthesia. Their result revealed six genes - COL4A1, ITGA2, MYO10, ROBO3, SLC9A6 and SLIT2, which caused hyperconnectivity of neuron during the early childhood.
| HGNA Symbol | Full Name | Exon Count | Location |
|---|---|---|---|
| COL4A1 | collagen type IV alpha 1 chain | 54 | chr 13 |
| ITGA2 | integrin subunit alpha2 | 30 | chr 5 |
| MYO10 | myosin X | 44 | chr 5 |
| ROBO3 | roundabout guidance receptor 3 | 29 | chr 11 |
| SLC9A6 | solute carrier family 9 member A6 | 20 | chr X |
| SLIT2 | slit guidance ligand 2 | 40 | chr 4 |
Choose SLIT2 for its low exon number
We then use NCBI GeneBank for more information of SLIT2.
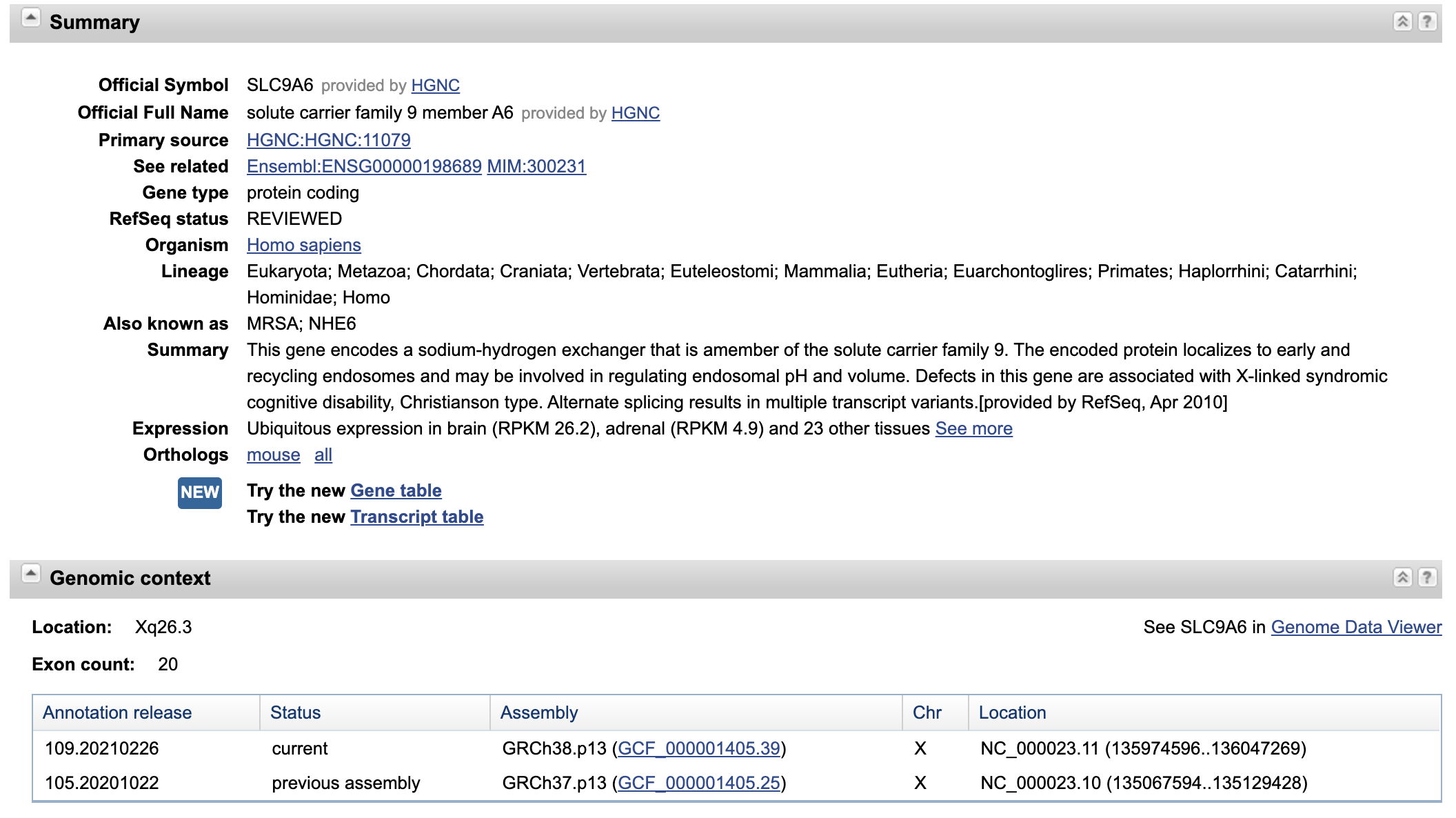 we can see a lots of information about Gene SLIT2. From this point, we will begin to feel overwhelmingly about the data
around the molecular biology. And this is just a tip of iceberg. SLC9A6 is a gene encoding a solute proteins regulating pH and volumn of endosomal.
It have also some alternative names such as MRSA(cool, just a abbrebiation of a super bug), NHE6. When we scrolling down the page, we can see more
details on the sequence of the gene and its genomic structure.
we can see a lots of information about Gene SLIT2. From this point, we will begin to feel overwhelmingly about the data
around the molecular biology. And this is just a tip of iceberg. SLC9A6 is a gene encoding a solute proteins regulating pH and volumn of endosomal.
It have also some alternative names such as MRSA(cool, just a abbrebiation of a super bug), NHE6. When we scrolling down the page, we can see more
details on the sequence of the gene and its genomic structure.
In the begining, we will feel the tracks of genome browseer is not so related to one's purpose, we can further configure the track and
only shows the track related to what we want to do. There is a button 'Tracks' upper right site of the genome browser, which enable you
to configure the tracks you want.
We may not want to edit this genes and let someone or oragnism got sick, so we need to see more about the mutation on this gene cause disease and
avoid this region for editing.
Woop! We found that lots of disease study unable to load and have no such information, which also show the data harmonization and
we still have lots of space to dig it for new knowledge discovery.
Because the disease related project have no related information on this genes or .... some bugs from the NCBI, we can only
see the variant level information on this genes.We should probably avoid this region for editing, bacause of the pathogenic possibility.
Furthermore, we can see this protein expression specifically in the brain tissue, which seems to make sense that them relate to color-sound Synesthesia.
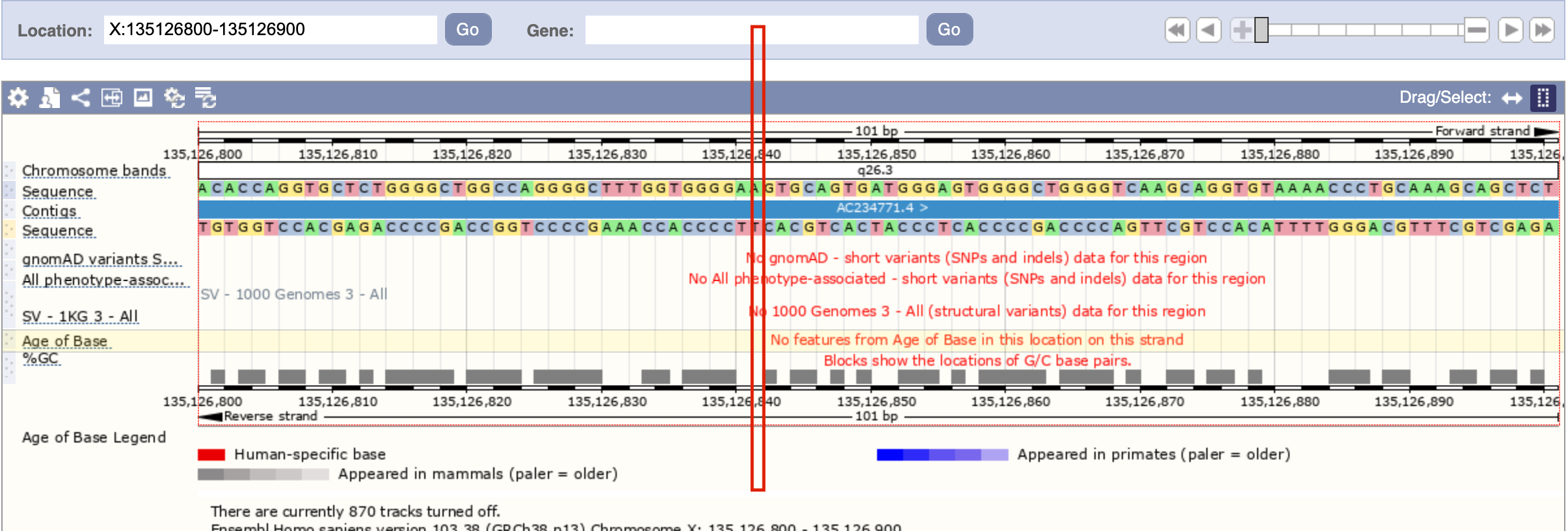
From PNAS publication we mentioned, the SLC9A6 varaint observed in the members of synesthesia family is located at X:135126891, A/T change. This region
is related to 3' UTR without known pathogenic (this is great!). The below is the sequence around the X:135126891, and the red box hightlight the position
we want to modify and make the sensory ability elevation. This region seems to have no known pathogenic variants.
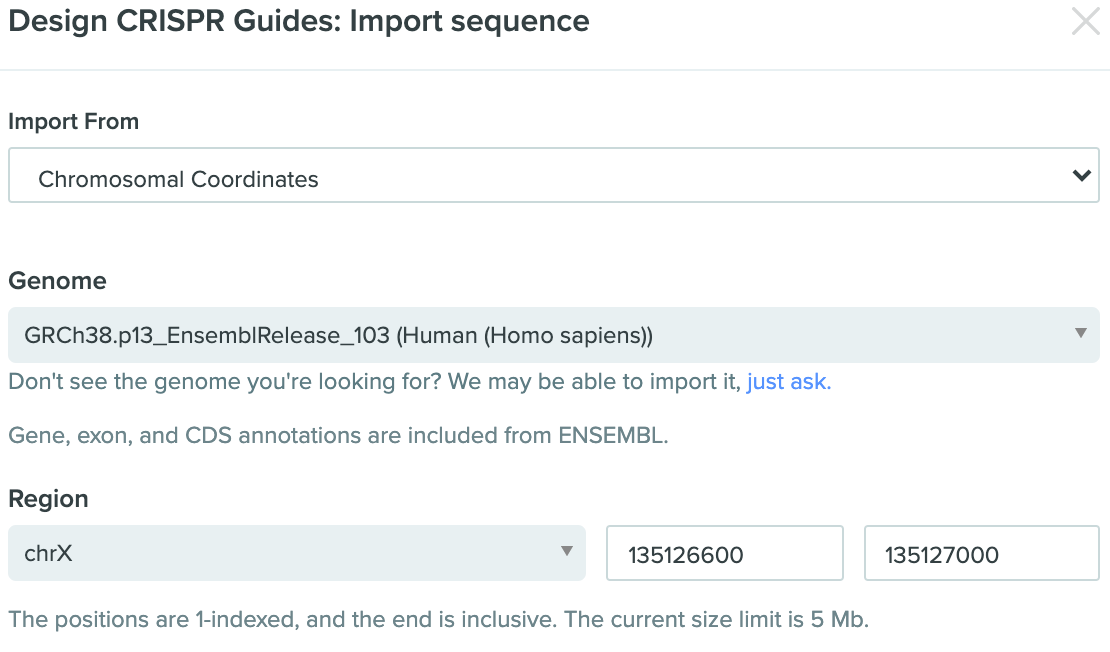
Using Benchling for CRISPR design
Benchling is really great web app for synthetic biology experiment deisgn, it provide free service for academic usage. we use Benchling for the CRISPR design. First, we choose to import a sequence from chromosome coordinate rather then gene Because the position we want to edit is at the 3' UTR, may not show at the ENSG data.
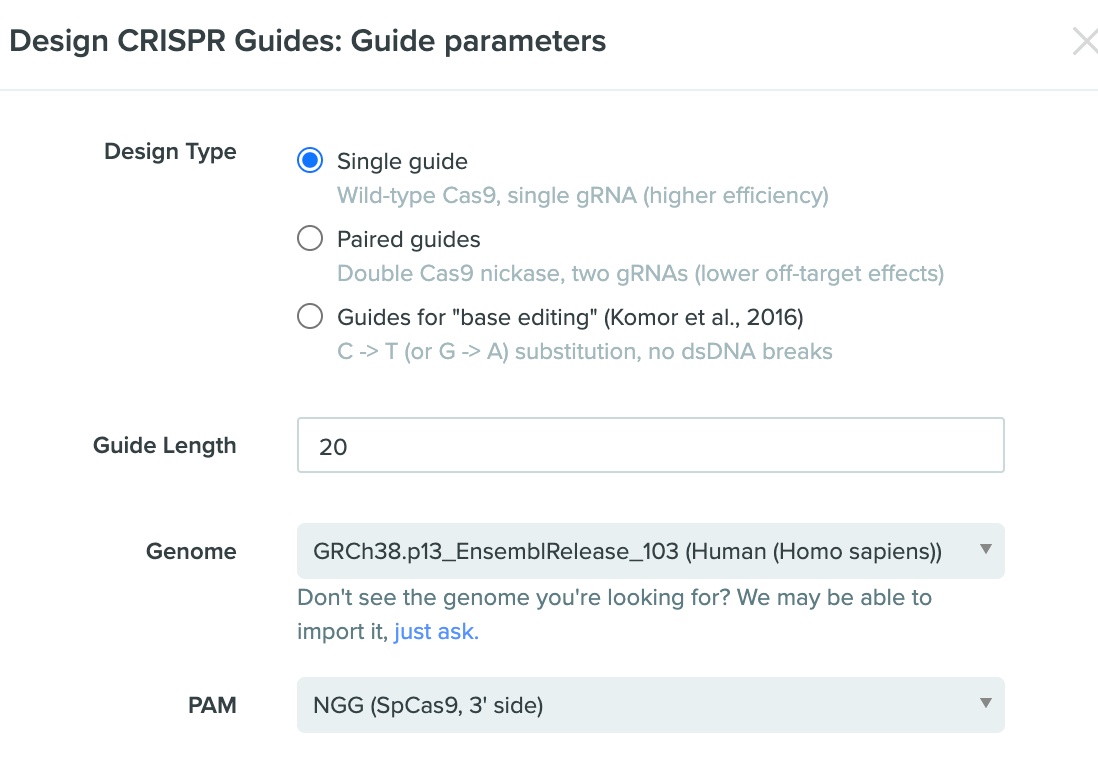
After you choose the methods to import the sequence, the Benchling will ask you what kinds of CRISPR guide you want. For a rookie on this stuff, I choose the single guide CAS9, seems to be more... simpler.
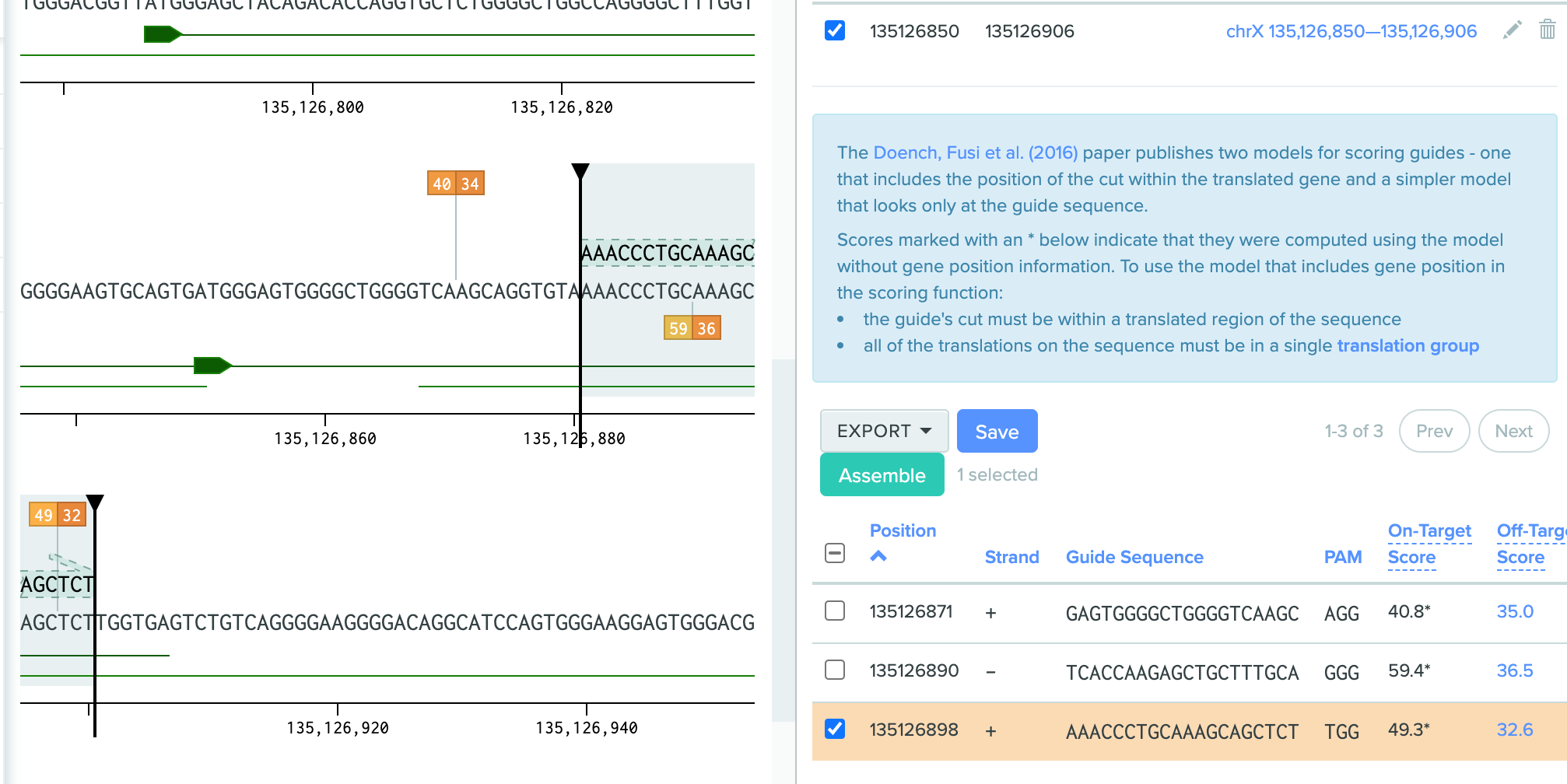
We can then see the hot spot where locate the PAM sequence and allow us to design editing. And I choose the one overlap the position at chrX:135126891 .Due to the human genome is double-strands, and SLC9A6 is located on forward. The gRNA located on the positive strands is what we look for.
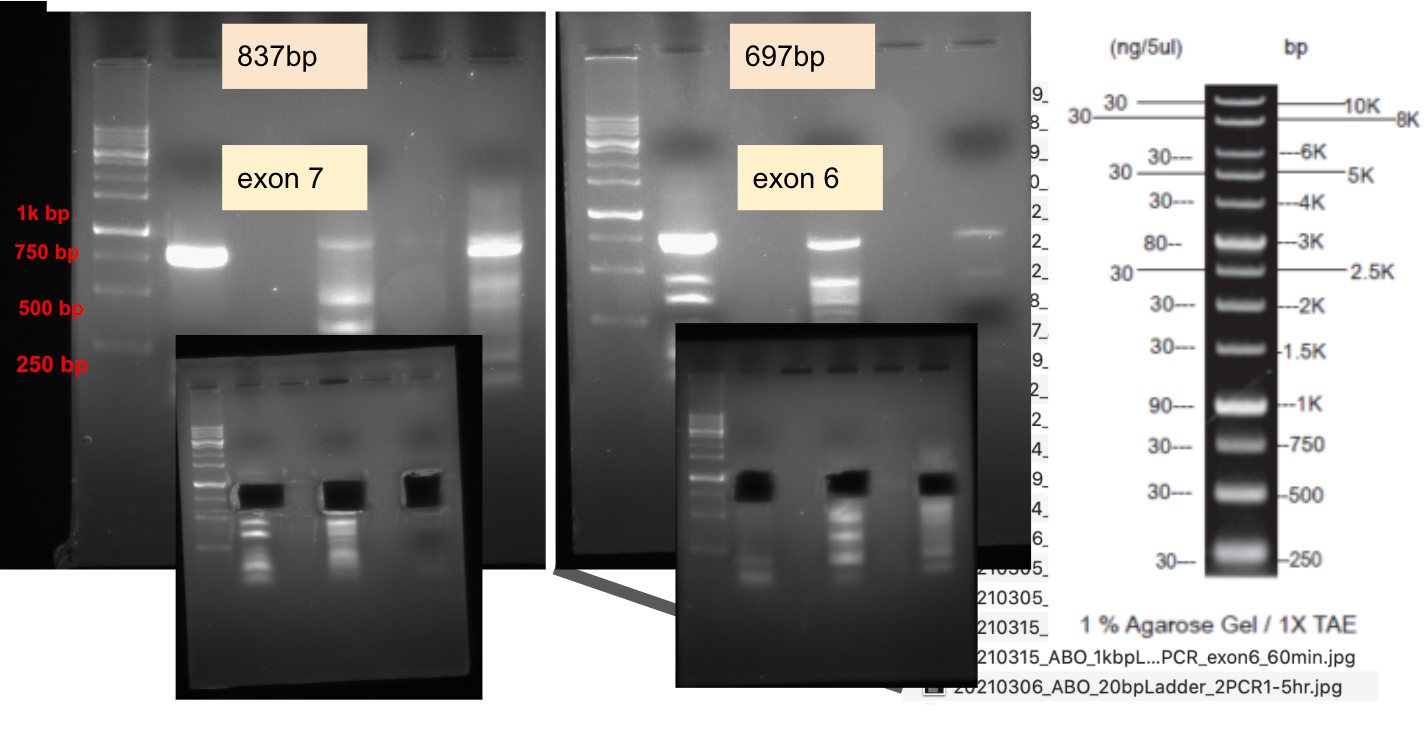
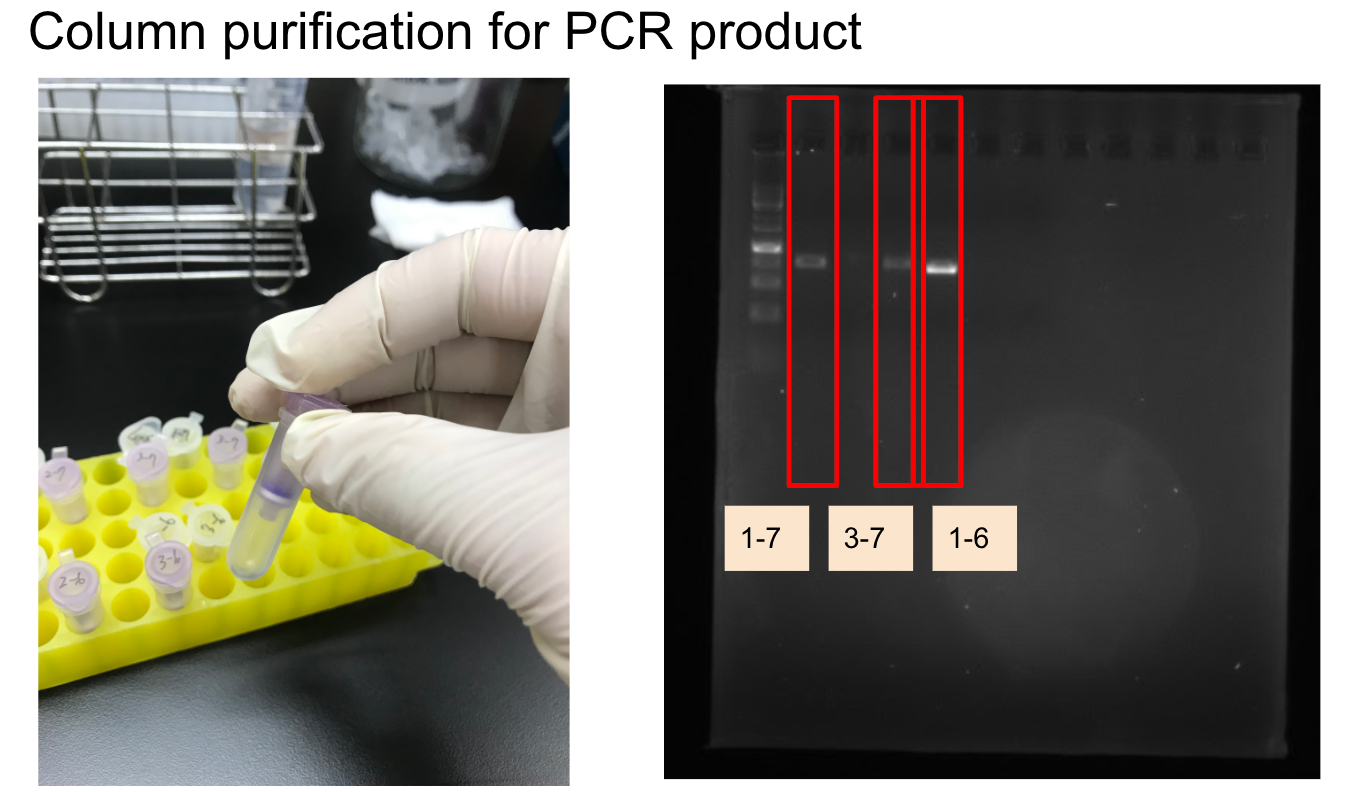
Lab Practice: typing ABO blood type with saliva DNA


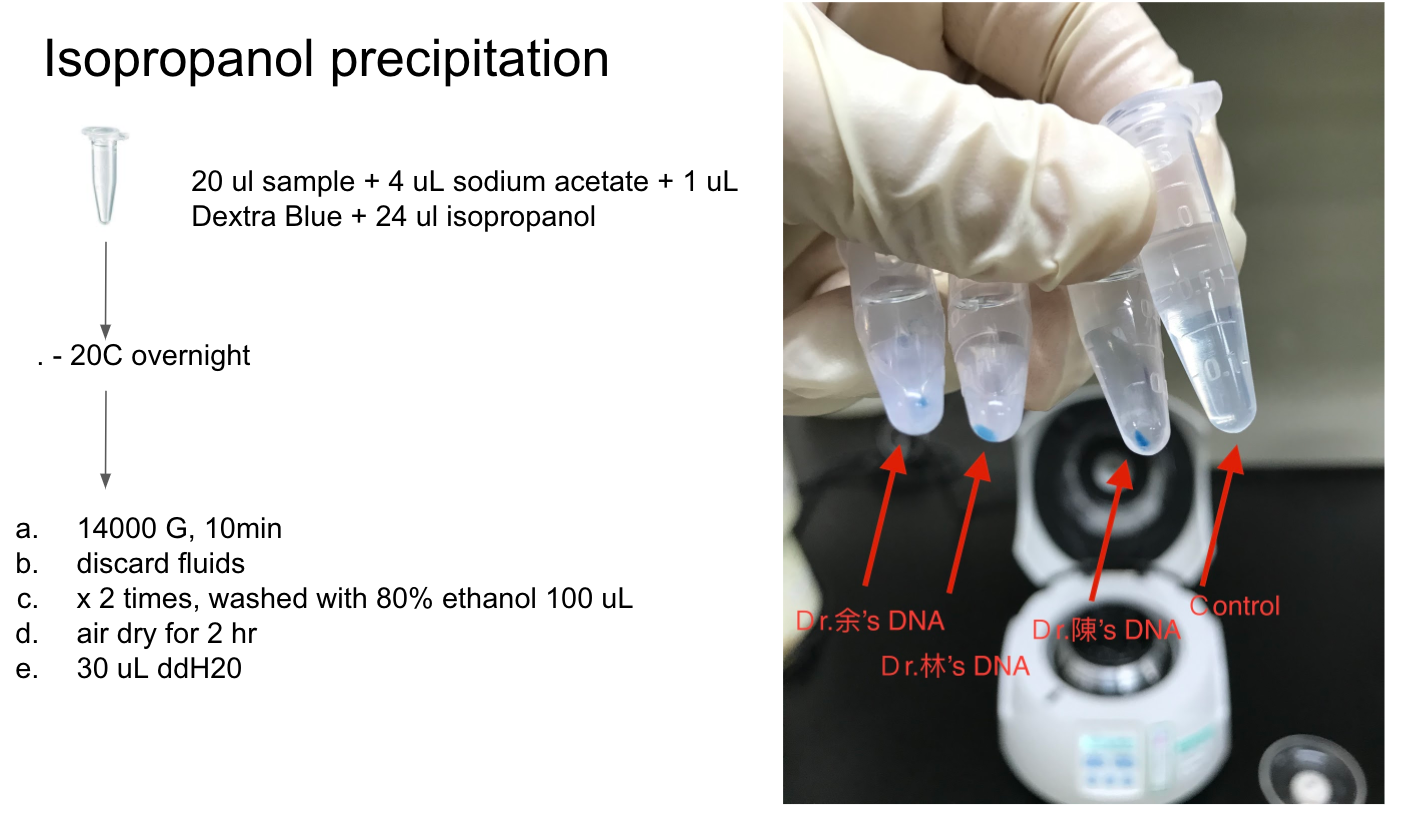
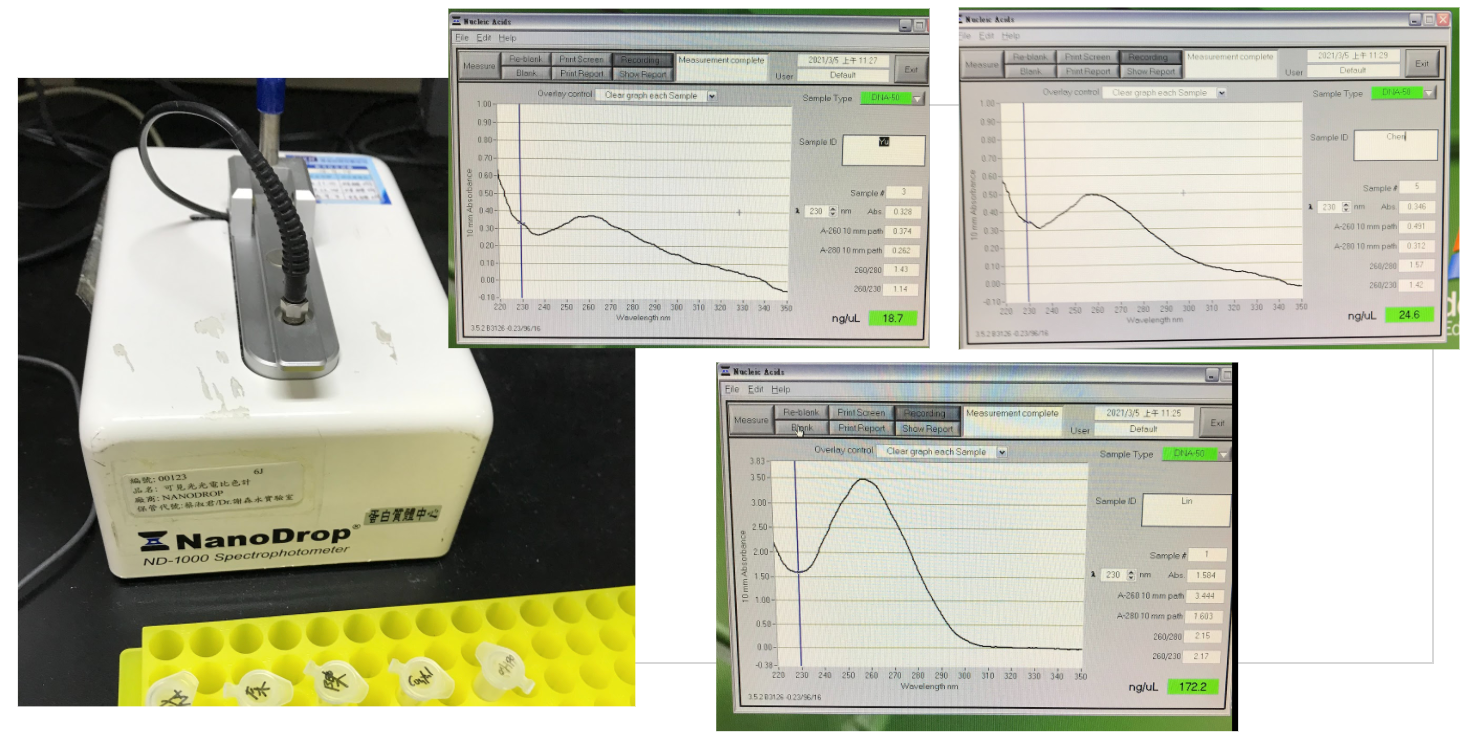
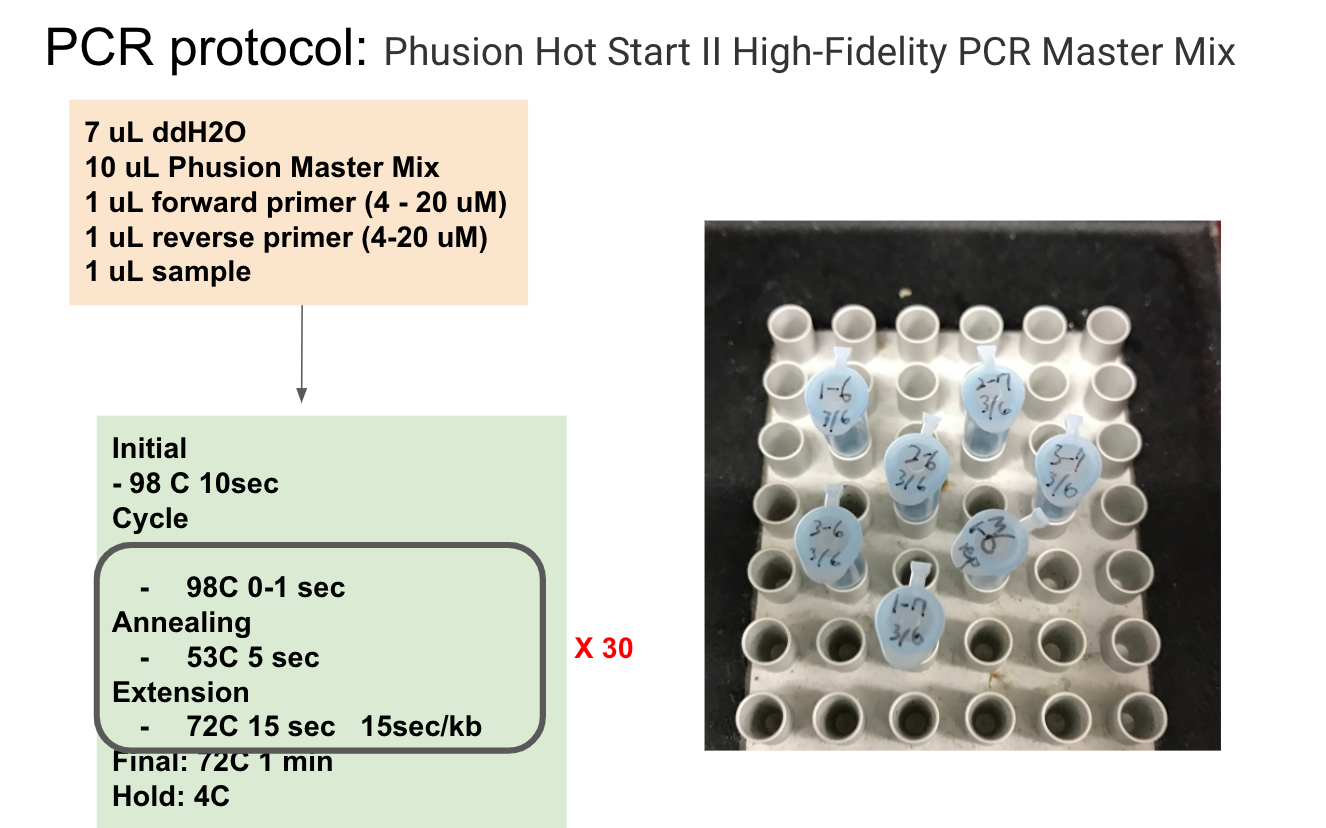

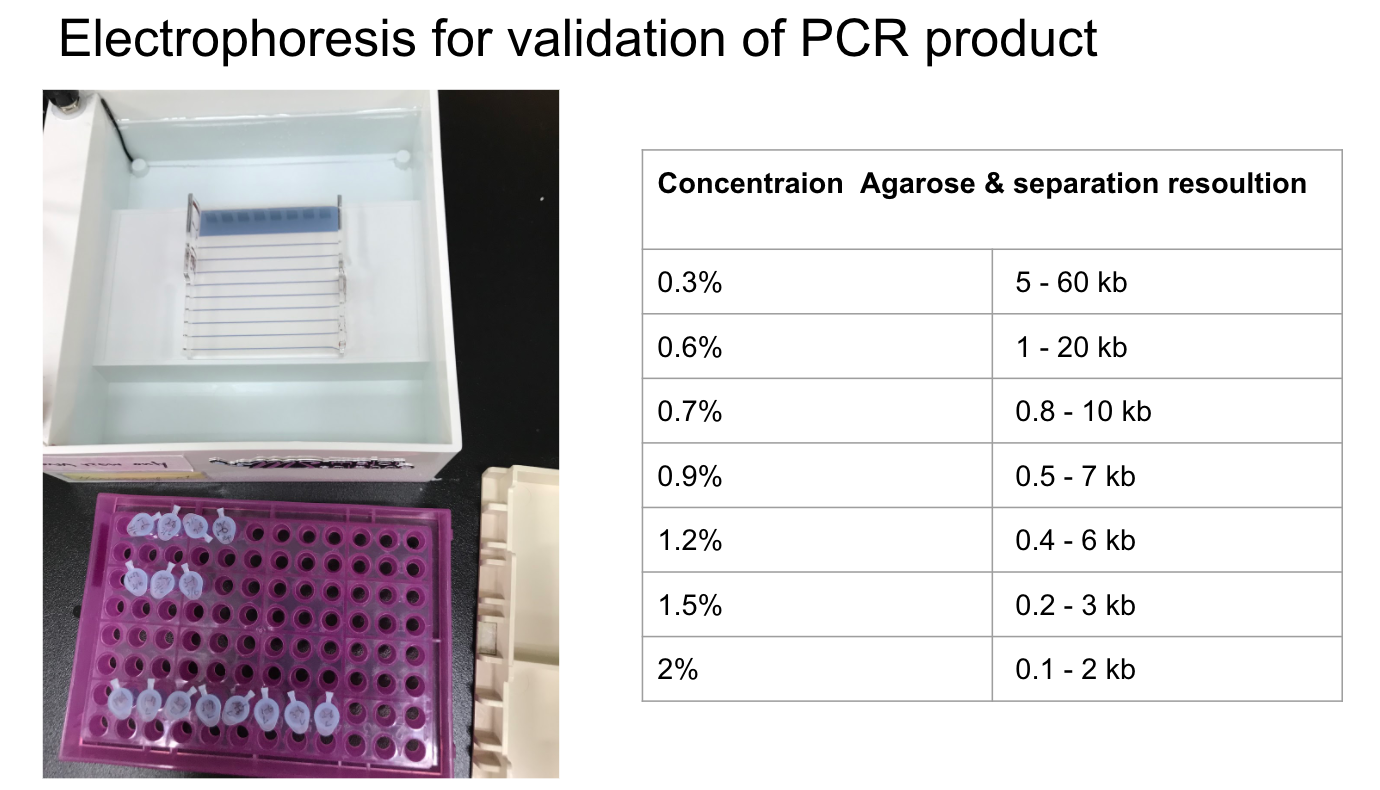
Reference
- Addgene. GRUSPR Overview: https://www.addgne.org/guides/crispr/
- 2011. Survival of the synesthesia Gene: Why DO People Hear Colors and Taste Words?. PLos Biology
- 2018. Rare variants in axonogenesis genes connect three families with sound-color synesthesia, PNAS
- 2013. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Treands Biotechnology
- 2016. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnology
- 2019. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature